Journal of Animal Research: v.10 n.2, p. 231-236. April 2020
DOI: 10.30954/2277-940X.02.2020.10
Karyomorphological Studies of Kangayam, Pulikulam, Crossbred Jersey and Crossbred Holstein Friesian Bulls
ABSTRACT
The present study was carried out in four genetic groups of cattle, viz. Kangayam, Pulikulam, crossbred Jersey and crossbred Holstein Friesian, to compare the karyomorphological pattern between Bos indicus and Bos taurus x indicus bull calves. Metaphase chromosomal spreads obtained by short term lymphocyte culture technique revealed chromosomal complement (2n) of 60, with 29 pairs of autosomes and one pair of sex chromosomes in four groups. All the autosomes were acrocentric, X-chromosome was sub-metacentric and Y-chromosome was acrocentric in Bos indicus and metacentric in crossbred bulls. There was no significant difference in relative length, arm ratio, centromeric index and morphological index of autosomes and X-chromosome between indicine and taurine groups; but Y-chromosome differed significantly (P˂0.01) in relative length between Bos indicus and Bos taurus x indicus crosses. Y-chromosome polymorphism could help in the determination of breed origin and male lines used in the breeding programmes in order to prevent the possible interferences in the process of reproduction.
Keywords: Crossbreds, Kangayam, Pulikulam, relative length, Y-chromosome polymorphism
Among the breeding strategies, crossing of non-descript cows with exotic bulls has gained more importance so as to increase the milk production of non-descript cows in India. According to 20th Livestock Census (2019), the indigenous/non-descript cattle population in India is 142.11 millions, while the exotic/crossbred cattle population is 50.42 million accounting for 26.19 per cent of total cattle population. This has increased by 26.9% compared to previous census (39.73 millions). This is achieved by extensive coverage of crossbred and non- descript cows through artificial insemination network, utilizing the semen of elite sires.
How to cite this article: Vemula, H., Periya, K., Doraiswamy, C. and Alagappan, M.G. (2020). Karyomorphological studies of kangayam, pulikulam, crossbred jersey and crossbred holstein friesian bulls. J. Anim. Res., 10(2): 231-236.
The sires selected for frozen semen production are screened for chromosomal polymorphism, since chromosomal incompatibility results in fertility related problems. Though the chromosomal abnormalities are rare in mammalian species, Y-chromosome polymorphism (Stranzinger et al., 2007; Wurtser and Benirschke, 1960) had been reported among sex chromosomes, while C-band polymorphism observed in autosomes (Switonski et al., 1983; Switonski, 1984). Karyologically, the morphology of autosomes and X-chromosome is similar in Bos taurus and Bos indicus, whereas Y-chromosome exhibits differences (Parada et al., 2018). During meiosis, the synaptonemal complex is formed between short homologues of pseudo-autosomal regions (PAR) between X- and Y-chromosomes. This exchange between sex chromosomes will alter the length of the pseudo-autosomal region, thereby changing the arm ratio (Ohno and Weiler, 1962). In addition, it was also stated by Pearson and Borrow (Pearson and Borrow, 1970), if pairing of X and Y was not possible, then meiosis would be stopped and sperm production would not occur. Previously there were reports of reproductive failures in crossbreds of Bos taurus and Bos indicus (Rendel, 1980). Structural and numerical chromosomal abnormalities may not be the reasons for the fertility related problems; but, minor differences in the morphometric measurements such as relative length and arm ratio would cause reproductive failures (Stranzinger et al., 2007). With this backdrop, the current study was conducted to compare the morphometric measurements of Kangayam, Pulikulam, Jersey crossbred and Holstein Friesian crossbred, to find chromosomal differences among the Bos indicus and Bos taurus x Bos indicus cattle.
MATERIALS AND METHODS
A total of 122 bull calves (22 Kangayam, 13 Pulikulam, 48 crossbred Jersey and 39 crossbred Holstein Friesian) produced through field progeny testing programme were screened cytogenetically in the Department of Animal Genetics and Breeding, Madras Veterinary College, Chennai, India between October, 2018 to July 2019. Metaphase chromosomes of the peripheral blood lymphocytes were prepared through short term lymphocyte culture technique according to Bharti et al. (2017). Blood lymphocytes were cultured by adding 7 ml of complete chromosomal medium (Euroclone, Italy) to 0.5 ml of whole blood. The culture was incubated for 72 hours at 37ºC and added 8 µg of colchicine one and half hour prior to harvesting the culture. Later, the cells were treated with hypotonic solution (0.075M KCl) for 30 minutes at 37ºC and then fixation in carnoy’s solution (3:1 ratio of Methanol : Glacial acetic acid). Cell suspension was dropped onto chilled wet slides from 2 to 3 feet height with pressure. Slides were air dried, stained with 5 per cent Giemsa stain for 30 minutes. Slides were screened under microscope and good metaphase spreads were photographed. Karyotyping was done with applied spectral imaging software Olympus, USA) as per the standard nomenclature (Di Berardino et al. 1989; Di Berardino et al., 2000). The length of the chromosomes was measured in millimeter with an accuracy of 0.05 mm, using a vernier calliper and morphometric measurements were recorded. Data on morphometric measurements were subjected to one-way ANOVA using IBM SPSS Statistics 20 to observe the significant differences between the genetic groups.
Formulas for the calculation of morphometric measurements were given below:

RESULTS AND DISCUSSION
Modal chromosome number and morphology
The bull calves of both Bos indicus and Bos taurus x indicus crosses analyzed in this study revealed a modal chromosome number (2n) of 60. The metaphase spreads and respective karyotypes of indigenous and crossbred bulls are presented in Fig. 1 and 2 respectively. All the autosomes were acrocentric, X -chromosome was sub- metacentric and Y -chromosome was acrocentric in Bos indicus and metacentric in crossbred bulls. These findings were in accordance with the earlier observations made in Bos indicus breeds of cattle viz. Khillari (Nakod, 2012), Sahiwal (Subramanyam, 2013), Tho-Tho (Choudhury et al., 2014), Punganur (Bharathi et al., 2015), Malnad Gida (Suresh et al., 2015), Ongole (Bharti et al., 2017), Alambadi (Parameswari et al., 2019), dwarf breeds of India (Rajesh Kumar et al., 2016) and Indonesian breeds (Ciptadi et al., 2017); Bos taurus breed, Holstein Friesian (Jamir et al., 2015) and Bos indicus x Bos taurus crosses viz. Jersey and Holstein Friesian crossbreds (Subramanyam, 2013; Nagpure et al., 2001; Cauveri and Sivaselvam, 2015).
There were two hypotheses given by the researchers for the differences in Y-chromosome morphology between taurine and indicine bulls. First being translocation of chromosomal arms between autosomes and Y-chromosome (Rendel, 1980) which caused a balanced stable chromosomal polymorphism as evidenced from lower calving rates in F2 generation of Brahman crosses with Bos taurus. Second hypothesis was centromere transposition or pericentric inversion which might be the reason for morphological differences in Y-chromosome (Goldammer et al., 1997; Di Meo et al., 2005). Comparative banding, in situ hybridization and southern blotting with specific DNA probes revealed pericentric inversion occurred in Y-chromosome between Bos taurus and Bos indicus cattle (Goldammer et al., 1997).
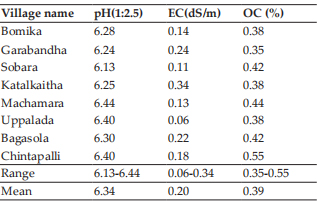
Fig. 1: Metaphase spread and karyotype of Kangayam bull calf
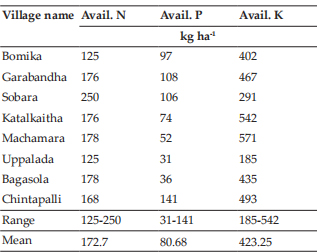
Fig. 2: Metaphase spread and karyotype of crossbred Jersey bull calf
Relative length
Analysis of variance revealed that there was no significant difference in relative lengths of autosomes and X-chromosome between the indigenous and crossbred bulls which concurred with the earlier studies in Sahiwal and Jersey crossbreds (Subramanyam, 2013), Punganur (Bharathi et al., 2015) and Malnad Gidda (Suresh et al., 2015). But, there was a significant difference (P<0.01) in relative length of Y-chromosome between Bos indicus and Bos taurus × indicus bulls (Stranzinger et al., 2007; Bharathi et al., 2015). The significant difference in the relative length of Y-chromosome was attributed to pericentric inversion causing loss of genetic material (Teale et al., 1995). The pericentric inversion should have resulted from splitting and dislocation of heterochromatin region and the influence of PAR region as opined by Stranzinger et al. (2007). In general, the relative length of a chromosome indicates the contribution of concerned chromosome to the haploid genome of the species under investigation, since it is important for classification of chromosomes, identification of deletions, duplications and translocations (Hansen, 1980). The relative lengths of X- and Y-chromosomes did not differ significantly between Kangayam and Pullikulam breeds.
The means of relative length of chromosomes in various genetic groups of cattle were given in Table 1. The relative lengths of autosomes were ranging from 5.30 ± 0.06 (1st chromosome) to 1.72 ± 0.04 per cent (29th chromosome); and 5.27 ± 0.07 (1st chromosome) to 1.70 ± 0.05 per cent (29th chromosome) in Kangayam and pulikulam bulls respectively. These values were similar to the earlier findings in indigenous breeds such as Red Kandhari (Katkade, 2005), Deoni (Balaji et al., 2006), Sahiwal (Subramanyam, 2013), Ongole (Bharti et al., 2017) and Alambadi (Parameswari et al., 2019).
The mean relative length of autosomes ranged from 5.36 ± 0.06 (1st chromosome) to 1.66 ± 0.04 (29th chromosome) in crossbred Jersey and 5.20 ± 1.73 (1st chromosome) to 1.73 ± 0.03 (29th chromosome) in crossbred Holstein Friesian bulls. The values for X- and Y-chromosomes were 5.83 ± 0.07 and 1.78 ± 0.06 in crossbred Jersey; and 5.50 ± 0.05 and 1.96 ± 0.06 in crossbred Holstein Friesian bulls respectively. These observations were slightly different from values reported in Holstein Friesian in Maharashtra (Nagpure et al., 2001), and Jersey crossbred cattle in Andhra Pradesh and Karnataka, India (Subramanyam et al., 2013; Suresh et al., 2015). Among the whole chromosomal complement, X-chromosome was the longest and Y was the smallest.
Table 1: Mean relative length of autosomes and allosomes in Kangayam, Pulikulam, crossbred Jersey and crossbred HF bull calves
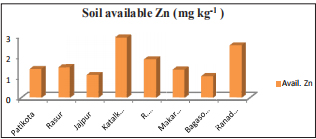
** Significant (P<0.01); NS: Non - significant; Means bearing different superscripts differ significantly (P<0.01).
Arm ratio
The magnitude of the arm ratio indicates the nature of the chromosome, and alterations in the arm ratio denotes structural variations in the concerned chromosome pair (29). In the present study, the means for arm ratio for X -chromosome in Kangayam, Pulikulam, crossbred Jersey and crossbred Holstein Friesian bulls were 2.14 ± 0.08, 2.06 ± 0.10, 2.11 ± 0.04 and 2.16 ± 0.07, respectively. The mean arm ratios of Y-chromosome was 1.18 ± 0.03 in crossbred Jersey and 1.16 ± 0.03 in crossbred Holstein Friesian. The values found in the present study were in agreement with the earlier reports in Deoni (Balaji et al., 2006); Kangayam (Kumarasamy et al., 2008); Umblachery (Kumarasamy et al., 2006); Sahiwal and Jersey crossbreds (Subramanyam, 2013); Punganur (Bharathi et al., 2015); Malnad Gidda (Suresh et al., 2015); Ongole (Bharti et al., 2017) and Alambadi (Parameswari et al., 2019). However, all these findings confirmed the sub-metacentric nature of X-chromosome in indigenous and crossbred bulls; and metacentric nature of the Y-chromosome in crossbred Jersey and Holstein cattle.
Centromeric Index
Centromeric index is a morphological characteristic of the bi-armed chromosomes which determines the location of the centromere. Chromosomes were identified in pairs based on the relative length and position of the centromere (Harshini, 2017). The mean centromeric indices of X-chromosome were 0.32 ± 0.007, 0.34 ± 0.011, 0.35 ± 0.004 and 0.32 ± 0.006 in Kangayam, Pulikulam, crossbred Jersey and crossbred Holstein bulls, respectively, while the means of Y-chromosome were 0.46 ± 0.007 and 0.46 ± 0.006 in crossbred Jersey and crossbred Holstein Friesian respectively. These values indicate the sub-metacentric nature of the X -chromosome in cattle, as the centromere was little away from the midpoint, and metacentric nature of the Y-chromosome in crossbred cattle due to midpoint positioning of the centromere. These values were in agreement with the findings of earlier researchers compared.
Morphological index
The morphological index is directly proportional to the total length of the chromosome. Hence, change in the length of the chromosome results in variation in morphologicl index. The means for morphological index of X-chromosome were 4.20 ± 0.18, 4.22 ± 0.31, 4.56 ± 0.27 and 4.38 ± 0.15 in Kangayam, Pulikulam, crossbred Jersey and crossbred Holstein bulls, respectively; while, the values for Y-chromosome were 3.48 ± 0.1 and 3.57 ± 0.2 in crossbred Jersey and Holstein bulls. These values concurred with the earlier studies on Deoni (Balaji et al., 2006), Sahiwal and Jersey crossbreds (Subramanyam, 2013), Punganur (Bharathi et al., 2015), Ongole (Bharti et al., 2017) and Alambadi (Parameswari et al., 2019) breeds of cattle.
CONCLUSION
Chromosomal number and morphology are the primary requirements for thorough understanding of the genetics of a species and any deviation from such as nombre fundamentale (NF) would pave the way for the determination of fertility related problems. In this study, the crosses have been made between taurine and indicine breeds which are classified as two different sub-species of cattle, and investigating the chromosomal structural variations becomes mandatory to make decisions on selection for semen production thereby preventing the possible interference in the process of reproduction. From the results obtained, it could be concluded that the sires chosen for frozen semen production are devoid of any chromosomal abnormality, suggestive of adoption of sound breeding strategies in selecting high genetic merit bulls.
ACKNOWLEDGEMENTS
The financial support research grant provided by Department of Animal Genetics and Breeding, Madras Veterinary College, Chennai is gratefully acknowledged.
REFERENCES
Bharti, A., Reddy, P.P., Gnana, P.M. and Sakaram, D. 2017. Cytogenetic characterization of Ongole cattle. Int. J. Adv. Biol. Res., 7(3): 574-577.
Bhatia, S. and Shanker, V. (1991). Cytogenetic analysis of Gaddi goats. Indian J. Anim. Sci., 61: 646-648.
Balaji, R., Gupta, B.R., Rao, G.N. and Reddy, G.V. 2006. Cytogenetic characterization of Deoni cattle. Indian J. Anim. Res., 40(1): 20-24.
Bharathi, G., Sakaram, D., Gnana Prakash, M. and Ramesh Gupta, B. 2015. Cytogenetic characterization of Punganur cattle. Int. J. Sci. Appl. Res., 2: 46-52.
Cauveri, D. and Sivaselvam, S.N. Karyotyping to identify indigenous male parent-age in crossbred calves. Int. J. Adv. Res., 3(12): 853-855.
Choudhury, H., Sarmah, V.B., Dhali, A., Choudhury, M.D. and Vidyarthi, V.K. 2014. Chromosomal characteristics of Tho- Tho cattle (Bos indicus) in Nagaland, India. Afr. J. Agric. Res., 9: 2894-2900.
Ciptadi, G., Ihsan, M.N., Rahayu, S,, Budiarto, A. and Wahjuningsih, S. 2017. The comparison of goat chromosome analysis result using simple G banding by manual and software cytovision image analysis. Res. J. Life Sci., 4(2): 106-110.
Di Berardino, D., Hayes, H., Fries, R. and Long, S. (eds). 1989. ISCNDA. International System for Cytogenetic Nomenclature of Domestic Animals. Cytogenet. Genome Res., 53 (2-3): 65-79.
Di Berardino, D., Di Meo, G.P., Gallagher, D.S., Hayes, H. and Iannuzzi, L. (coordinator) (eds). 2000. ISCNDB. International System for Chromosome Nomenclature of Domestic Bovids. Cytogenet. Cell Genet., 92 (3-4): 283.
Di Meo, G.P., Perucatti, A., Floriot, S., Incarnato, D. and Rullo, R. 2005. Chromosome evolution and improved cytogenetic maps of the Y chromosome in cattle, Zebu, River buffalo, sheep and goat. Chromosome Res., 13: 349-355.
Goldammer, T., Brunner, R.M. and Schwerin, M. 1997. Comparative analysis of Y-Chromosome structure in B. taurus and B. indicus by FISH using region-specific, microdissected, and locus-specific DNA probes. Cytogenet. Cell Genet., 77: 238-241.
Hansen, K.M. 1980. The relative length of pig chromosomes, and a suggestion for a karyotype system. Ann. Genet. Sél. Anim., 12(4): 1.
Harshini, V. 2017. A comparative study on cytogenetic profile of crossbred and indigenous pigs. doctoral dissertation, Sri Venkateswara Veterinary University, Tirupati-517 502. India.
Jamir, J., Mukhopadyay, C.S., Arora, J.S., Brah, G.S., Cheema, R. and Kaur, M. 2015. Screening of dairy breeding bulls for chromosomal profile and its andrological attributes. Indian J Anim. Res., 49(1): 8-13.
Katkade, B.S. 2005. Cytogenetic Studies on Indian Domestic Cattle (Bos indicus). Doctoral dissertation, MAFSU, Nagpur, India.
Kumarasamy, P., Sivaselvam, S.N., Rajendran, R., Thangaraju, P. and Nainar, A.M. 2006. Chromosomal characterization of Umblachery breed of cattle (Bos indicus)–a famous South Indian breed of Tamilnadu, India. Indian J. Sci. Tech., 1(6): 1-3.
Kumarasamy, P., Sivaselvam, S.N., Thara, S., Thangaraju, P. and Nainar, A.M. 2008. Chromosomal studies on Kangayam cattle. Indian Vet. J., 83: 1072.
Nagpure, N.S., Pawde, A.M. and Koul, G.L. 2001. Karyological studies in Hariana, Holstein Friesian and their crossbreeds. Indian J. Anim. Sci., 71 (6): 551-555.
Nakod, N. 2013. Karyological Evaluation Of Khillar Cattle. Doctoral dissertation, MAFSU, Nagpur, India.
Ohno, S. and Weiler, C. 1962. Relationship between large Y-chromosome and side-by-side pairing of the XY-bivalent observed in the Chinese hamster, Cricetus Griseus. Chromosoma, 13 (1): 106-110.
Parada, R., Kawka, M., Sacharczuk, M., Urbański, P. and Jaszczak, K. 2018. Cytogenetic and genetic study of a Y-linked microsatellite polymorphism in Polish Black-and- White cattle breed. Saudi J. Biol. Sci., 25(7): 1406-1410.
Parameswari, S., Cauveri, D., Karthickeyan, S.M.K., Arunachalam, K. and Kumarasamy, P. 2019. Cytogenetic Characterisation of Alambadi Breed of Cattle in Tamil Nadu. Indian Vet. J., 96: 49-52.
Pearson, O.L. and Borrow, M. 1970. Definitive evidence for the short arms of the Y chromosome association with the X-chromosome during meiosis in the human male. Nature, 229: 959–960.
Rajesh, K.P., Rosaiah, K. and Nagaraju, N.S. 2016. Cytogenetic investigation of rare cattle breeds of India. J. chem. biol. phy. sci., 6(3): 571-575.
Rendel, J.M. 1980. Low calving rates in Brahman cross cattle. Theor. Appl. Genet., 58(5): 207-210.
Sahai, R. and Vijh, R.K. (1994). Cytogenetic techniques in farm animals-A Laboratory Manual, NBAGR Research Bulletin.
Stranzinger, G.F., Steiger, D., Kneubühler, J. and Hagger, C. 2007. Y chromosome polymorphism in various breeds of cattle (Bos taurus) in Switzerland. J. Appl. Genet., 48(3): 241-245.
Subramanyam, B. 2013. Cytogenetic characterization of Purebred and Crossbred Sahiwal cattle. Doctoral dissertation, Sri Venkateswara Veterinary University, Tirupati, India.
Suresh, S.C., Nagaraja, C.S. and Satheesha, G.M. 2015. Cytogenetic studies in Malnad Gidda cattle. Wayamba J. Anim. Sci., 1424965516: 1059-1065.
Switonski, M., Fries, R. and Stranzinger, G. 1983. C-band variants of telocentric chromosomes in swine: Evidence and inheritance studies. Génét. Sél. Evol., 15(4): 469.
Switonski, M. 1984. Cytogenetic examination of bulls. Variants of the Y chromosome. Genet. Pol., 25: 427.
Teale, A.J., Wambugu, J., Gwakisa, P.S., Stranzinger, G. and Bradley, D. 1995. A polymorphism in randomly amplified DNA that differentiates the Y-chromosomes of Bos indicus and Bos tasurus. Anim. Genet., 26: 243-248.
Wurster, D.H. and Benirschke, K. 1960. Chromosome studies in the super family Bovidae. Chromosoma, 25(2): 152-171.